Our Research
-
Gene regulation through non-coding RNAs in bacteria
-
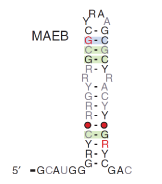 A lot of recent discoveries have shown that RNA has a very wide diversity of function. Furthermore, many more are expected to be found and a lot of research is still needed to find them and better understand their mechanisms.
A lot of recent discoveries have shown that RNA has a very wide diversity of function. Furthermore, many more are expected to be found and a lot of research is still needed to find them and better understand their mechanisms. -
 We use computational tools to find new RNAs and better understand them, as well as tools from biochemistry, molecular biology and genetics. Some of our model organisms include Pseudomonas and Burkholderia species, bacteria that are widespread in the environment, but can sometimes cause serious infections as opportunistic pathogens. We are also interested in finding ncRNAs from bacterial communities in various environments to help us better understand how these communities interact with their environment.
We use computational tools to find new RNAs and better understand them, as well as tools from biochemistry, molecular biology and genetics. Some of our model organisms include Pseudomonas and Burkholderia species, bacteria that are widespread in the environment, but can sometimes cause serious infections as opportunistic pathogens. We are also interested in finding ncRNAs from bacterial communities in various environments to help us better understand how these communities interact with their environment. -
Metabolic engineering and synthetic biology
- In parallel, we are interested in using our knowledge regarding ncRNAs to do genetic engineering for diverse applications. Notably, we use small RNAs to control gene expression to force bacteria to produce useful molecules towards a more "green" approach to supply the industry. In fact, we are collaborating with industrial partners that want to use bacteria as miniature factories for large scale production of chemicals.
-
Aptamers and biosensors
- Since synthetic aptamers can be selected against any target (metabolite, chemical contaminant, proteins or even whole cells) and then used as biosensors or "molecular targeting tools", we have many projects in collaboration where aptamers are conjugated with nanoparticles, optical fibers or other molecules such as peptides with therapeutic perspectives.
Our research is not limited to a few species. We are interested in all bacteria and archaea, which we compare with bioinformatic tools to find conserved RNAs. Thus, we are also interested in ncRNAs found in Mycobacterium tuberculosis which we study with collaborations.
Aptamers are nucleic acids capable of binding and recognizing specifically a given molecule. There are natural versions (riboswitches), but they can also be selected through an in-vitro accelerated evolution approach called SELEX. Aptamers are thus amenable to detect chosen molecules for numerous applications. An example includes the combination of RNA-aptamers with an expression system to produce artifical riboswitches, which can be used to make bacteria-biosensors.
